Tutorial 1b: Data frames¶
(c) 2017 Justin Bois. This work is licensed under a Creative Commons Attribution License CC-BY 4.0. All code contained herein is licensed under an MIT license.
This tutorial was generated from an Jupyter notebook. You can download the notebook here.
import numpy as np
import pandas as pd
import bokeh.io
import bokeh.plotting
bokeh.io.output_notebook()
In this tutorial, we will learn how to load data stored on disk into a Python data structure. We will use Pandas to read in CSV (comma separated value) files and store the results in the very handy Pandas DataFrame. This incredibly flexible and powerful data structure will be a centerpiece in the rest of this course and beyond. In this tutorial, we will learn about what a data frame is and how to use it.
The data set we will use comes from a fun paper about the adhesive properties of frog tongues. The reference is Kleinteich and Gorb, Tongue adhesion in the horned frog Ceratophrys sp., Sci. Rep., 4, 5225, 2014. You can download the paper here. You might also want to check out a New York Times feature on the paper here.
In this paper, the authors investigated various properties of the adhesive characteristics of the tongues of horned frogs when they strike prey. The authors had a striking pad connected to a cantilever to measure forces. They also used high speed cameras to capture the strike and record relevant data.
Importing modules¶
As I mentioned in the last tutorial, we need to import modules we need for data analysis. An important addition is Pandas, which we import as pd. As is good practice, I import everything at the top of the document.
with open('../data/frog_tongue_adhesion.csv', 'r') as f:
for _ in range(20):
print(next(f), end='')
The first lines all begin with # signs, signifying that they are comments and not data. They do give important information, though, such as the meaning of the ID data. The ID refers to which specific frog was tested.
Immediately after the comments, we have a row of comma-separated headers. This row sets the number of columns in this data set and labels the meaning of the columns. So, we see that the first column is the date of the experiment, the second column is the ID of the frog, the third is the trial number, and so on.
After this row, each row represents a single experiment where the frog struck the target.
CSV files are generally a good way to store data. Commas make better delimiters than white space (such as tabs) because they have no portability issues. Delimiter collision is avoided by putting data fields in double quotes when necessary. There are other good ways to store data, such as JSON, but we will almost exclusively use CSV files in this class.
Loading a data set¶
We will use pd.read_csv() to load the data set. The data are stored in a DataFrame, which is one of the data types that makes Pandas so convenient for use in data analysis. DataFrames offer mixed data types, including incomplete columns, and convenient slicing, among many, many other convenient features. We will use the DataFrame to look at the data, at the same time demonstrating some of the power of DataFrames. They are like spreadsheets, only a lot better.
We will now load the DataFrame.
# Use pd.read_csv() to read in the data and store in a DataFrame
df = pd.read_csv('../data/frog_tongue_adhesion.csv', comment='#')
Notice that we used the kwarg comment to specify that lines that begin with # are comments and are to be ignored. If you check out the doc string for pd.read_csv(), you will see there are lots of options for reading in the data.
Exploring the DataFrame¶
Let's jump right in and look at the contents of the DataFrame. We can look at the first several rows using the df.head() method.
# Look at the contents (first 5 rows)
df.head()
We see that the column headings were automatically assigned. Pandas also automatically defined the indices (names of the rows) as integers going up from zero. We could have defined the indices to be any of the columns of data.
To access a column of data, we use the following syntax.
# Slicing a column out of a DataFrame is achieved by using the column name
df['impact force (mN)']
The indexing of the rows is preserved, and we can see that we can easily extract all of the impact forces. Note, though, that pd.read_csv() interpreted the data to be integer (dtype = int64), so we may want to convert these to floats, which we do with the .astype() method.
# Use df.astype() method to convert it to a NumPy float64 data type.
df['impact force (mN)'] = df['impact force (mN)'].astype(float)
# Check that it worked
df['impact force (mN)'].dtype
Now let's say we only want the impact force of strikes above two Newtons, the big strikes. Pandas DataFrames can conveniently be sliced with Booleans. We first make an array of Booleans that are True when the condition of interest is met, and False elsewhere.
# Generate True/False array of rows for indexing
inds = df['impact force (mN)'] >= 2000.0
# Take a look
inds
We now have an array of Booleans that is the same length as the DataFrame itself. Now, we can use the .loc feature of a DataFrame to slice what we want out of the DataFrame.
# Slice out rows we want (big force rows)
df_big_force = df.loc[inds, :]
# Let's look at it; there will be only a few high-force values
df_big_force
Using .loc allows us to index by row and column. We chose all columns (using a colon, :), and put an array of Booleans for the rows. We get back a DataFrame containing the rows associated with True in the arrays of Booleans.
So now we only have the strikes of high force. Note, though, that the original indexing of rows was retained! In our new DataFrame with only the big force strikes, there is no index 3, for example.
# Executing the below will result in an exception
df_big_force.loc[3, 'impact force (mN)']
This might seem counterintuitive, but it is actually a good idea. Remember, indices do not have to be integers!
There is a way around this, though. We can use the iloc attribute of a DataFrame. This gives indexing with sequential integers.
# Using iloc enables indexing by the corresponding sequence of integers
df_big_force['impact force (mN)'].iloc[3]
Aside: Tidy data¶
In our DataFrame, the data are tidy. The concept of tidy data originally comes from development of databases, but has taken hold for more general data processing in recent years. We will spend most of next week talking about tidy data and how to work with it, including reading this paper and this paper by Hadley Wickham for a great discussion of this. Tidy data refers to data sets arranged in tabular form that have the following format.
- Each variable forms a column.
- Each observation forms a row.
- Each type of observational unit forms a separate table.
As we can see, Kleinteich and Korb generously provided us with tidy data. Much more on tidy data next week....
More data extraction¶
Given that the data are tidy and one row represents a single experiment, we might like to extract a row and see all of the measured quantities from a given strike. This is also very easy with DataFrames using .loc.
# Let's slice out experiment with index 42
df.loc[42, :]
Notice how clever the DataFrame is. We sliced a row out, and now the indices describing its elements are the column headings. This is a Pandas Series object.
Now, slicing out index 42 is not very meaningful because the indices are arbitrary. Instead, we might look at our lab notebook, and want to look at trial number 3 on May 27, 2013, of frog III. This is a more common, and indeed meaningful, use case.
# Set up Boolean slicing
date = df['date'] == '2013_05_27'
trial = df['trial number'] == 3
ID = df['ID'] == 'III'
# Slice out the row
df.loc[date & trial & ID, :]
The difference here is that the returned slice is a DataFrame and not a Series. We can easily make it a Series using iloc. Note that in most use cases, this is just a matter of convenience for viewing and nothing more.
df.loc[date & trial & ID, :].iloc[0]
Renaming columns¶
You may be annoyed with the rather lengthy syntax of access column names. I.e., if you were trying to access the ratio of the contact area with mucus to the contact area without mucus for trial number 3 on May 27, 2013, you would do the following.
# Set up criteria for our seach of the DataFrame
date = df['date'] == '2013_05_27'
trial = df['trial number'] == 3
ID = df['ID'] == 'III'
# When indexing DataFrames, use & for Boolean and (and | for or; ~ for not)
df.loc[date & trial & ID, 'contact area with mucus / contact area without mucus']
There are reasons to preserve the descriptive (and verbose) nature of the column headings. For example, everything is unambiguous, which is always a plus. Beyond that, many plotting packages, including HoloViews, which we will learn about next week, can automatically label axes based on headers in DataFrames. Nonetheless, you may want to use shorter names to save keystrokes. So, let's change the name of the columns as
'trial number' → 'trial'
'contact area with mucus / contact area without mucus' → 'ca_ratio'
DataFrames have a nice rename method to do this.
# Make a dictionary to rename columns
rename_dict = {'trial number' : 'trial',
'contact area with mucus / contact area without mucus' : 'ca_ratio'}
# Rename the columns
df = df.rename(columns=rename_dict)
# Try out our new column name
df.loc[date & trial & ID, 'ca_ratio']
Notice that we have introduced a new Python data structure, the dictionary. A dictionary is a collection of objects, each one with a key to use for indexing. For example, in rename_dict, we could get what we wanted to rename 'trial number'.
# Indexing of dictionaries looks syntactially similar to cols in DataFrames
rename_dict['trial number']
We can go on and on about indexing Pandas DataFrames, because there are many ways to do it. For much more on all of the clever ways you can access data and subsets thereof in DataFrames, see the Pandas docs on indexing.
Computing with DataFrames and adding columns¶
We have seen that DataFrames are convenient for organizing and selecting data, but we naturally want to compute with data. Furthermore, we might want to store the results of the computations as a new column in the DataFrame. As a simple example, let's say we want to have a column with the impact force in units of Newtons instead of milliNewtons. We can divide the impact force (mN) columns elementwise by 1000, just as with Numpy arrays.
# Add a new columns with impoact force in units of Newtons
df['impact force (N)'] = df['impact force (mN)'] / 1000
# Take a look
df.head()
Creation of the new column was achieve by assigning it to Pandas Series we calculated.
We can do other calculations on DataFrames besides elementwise calculation. For example, if we wanted the mean impact force in units of milliNewtons, we can do this....
df['impact force (mN)'].mean()
Actually, we can compute all sorts of useful summary statistics all at once about the DataFrame using the describe() method. This gives the count, mean, standard deviation, minimum, 25th percentile, median, 75th percentile, and maximum for each column.
df.describe()
Some values are meaningless, like the mean trial number, but much of the information is very useful.
Now, say we wanted to compute the mean impact force of each frog. We can combine Boolean indexing with looping to do that.
for frog in df['ID'].unique():
inds = df['ID'] == frog
mean_impact_force = df.loc[inds, 'impact force (mN)'].mean()
print('{0:s}: {1:.1f} mN'.format(frog, mean_impact_force))
Notice that we have used the .unique() method to get the unique entries in the ID column. We will see next week that we can use Pandas's groupby() function to do this much more cleanly and efficiently, but for now, you can appreciate your ability to pull out what data you need and compute with it.
Creating a DataFrame from scratch¶
Looking back at the header of the original data file, we see that there is information present in the header that we would like to have in our DataFrame. For example, it would be nice to know if each strike came from an adult or juvenile. Or what the snout-vent length was. Going toward the goal of including this in our DataFrame, we will first construct a new DataFrame containing information about each frog.
One way do create this new DataFrame is to first construct a dictionary with the respective fields.
data_dict = {'ID': ['I', 'II', 'III', 'IV'],
'age': ['adult', 'adult', 'juvenile', 'juvenile'],
'SVL (mm)': [63, 70, 28, 31],
'weight (g)': [63.1, 72.7, 12.7, 12.7],
'species': ['cross', 'cross', 'cranwelli', 'cranwelli']}
Now that we have this dictionary, we can convert it into a DataFrame by instantiating a pd.DataFrame class with it.
# Make it into a DataFrame
df_frog_info = pd.DataFrame(data=data_dict)
# Take a look
df_frog_info
Nice!
Sometimes the data sets are not small enough to construct a dictionary by hand. Oftentimes, we have a two-dimensional array of data that we want to make into a DataFrame. As an example, let's say we have a Numpy array that has two columns, one for the snout-vent length and one for the weight.
data = np.array([[63, 70, 28, 31], [63.1, 72.7, 12.7, 12.7]]).transpose()
# Verify that it's what we think it is
data
To make this into a DataFrame, we again create pd.DataFrame instance, but this time we also specify the column keyword argument.
df_demo = pd.DataFrame(data=data, columns=['SVL (mm)', 'weight (g)'])
# Take a look
df_demo
That also works. Generally, any two-dimensional Numpy array can be converted into a DataFrame in this way. You just need to supply column names.
Merging DataFrames¶
Our ultimate goal was to add that information to the main DataFrame, df, that we have been working with. We can do it using tools we have already learned. For each row in the DataFrame, we can add the relevant value in each column. Because this will not be the final way I recommend doing this, I will do these operations on a copy of df using the copy() method.
# Make a copy of df
df_copy = df.copy()
# Build each column
for col in df_frog_info.columns[df_frog_info.columns != 'ID']:
# Make a new column with empty values
df_copy[col] = np.empty(len(df_copy))
# Add in each entry, row by row
for i, r in df_copy.iterrows():
ind = df_frog_info['ID'] == r['ID']
df_copy.loc[i, col] = df_frog_info.loc[ind, col].iloc[0]
# Take a look at the updated DataFrame
df_copy.head()
Note that I used the iterrows() method of the df_copy data frame. This iterator gives an index (which I called i) and a row of a DataFrame (which I called r). This method, and the analogous one for iterating over columns, iteritems(), can be useful.
But this approach seems rather clunky. We could do it more concisely using list comprehensions (if you don't know what those are, don't worry), but a much better way to do it is to use Pandas's built-in merge() method. Called with all the default keyword arguments, this function finds a common columns between two DataFrames (in this case, the ID column), and then uses those columns to merge them, filling in values that match in the common column. This is exactly what we want.
df = df.merge(df_frog_info)
# Check it out!
df.head()
Boo-yah! I think this example of merging DataFrames highlights the power of using them in your data analysis.
Plotting how impact forces correlate with other metrics¶
Let's say for a moment that the impact forces and how they correlate with other measures about the impact are what we are most interested in. Let's make a scatter plot of adhesion forces versus impact forces.
# Set up the plot
p = bokeh.plotting.figure(plot_height=300,
plot_width=500,
x_axis_label='impact force (mN)',
y_axis_label='adhesive force (mN)')
# Add a scatter plot
p.circle(df['impact force (mN)'], df['adhesive force (mN)'])
bokeh.io.show(p)
We see some correlation. The stronger the impact force, the stronger the adhesive force, with some exceptions, of course.
Making subplots¶
I will use this as an opportunity to demonstrate how to make subplots in Bokeh. Let's say we want to plot the adhesive force, total contact area, impact time, and contact pressure against impact force. To lay these out as subplots, we make the respective bokeh.plotting.figure.Figure objects one by one and put them in a list. We then use the utilities from bokeh.layouts to make the subplots. Let's start by making the list of figure objects. It will help to write a quick function to generate a plot.
def scatter(df, x, y, p=None, color='#30a2da', plot_height=300, plot_width=500):
"""Populate a figure with a scatter plot."""
if p is None:
p = bokeh.plotting.figure(plot_height=plot_height,
plot_width=plot_width,
x_axis_label=x,
y_axis_label=y)
p.circle(df[x], df[y], color=color)
return p
Ok, now we can build a list of plots.
# Things we want to plot
cols = ['impact time (ms)',
'adhesive force (mN)',
'total contact area (mm2)',
'contact pressure (Pa)']
plots = []
for col in cols:
plots.append(scatter(df,
'impact force (mN)',
col,
plot_height=200,
plot_width=400))
Now, to line them up in a column, we can use the bokeh.layouts.column() function to generate a layout, which we can then show using bokeh.io.show().
bokeh.io.show(bokeh.layouts.column(plots))
There are a few things I'd like to clean up about this figure. First, all plots share the same x-axis, so I would like to link the axes. This is accomplished by setting the x_range attribute of the respective bokeh.plotting.figure.Figure objects. Furthermore, since all the x-axes are the same, we do not need to label them all.
for p in plots[:-1]:
# Set eacn plot's x_range to be that of the last plot
p.x_range = plots[-1].x_range
# Only have x_axis label on bottom plot
p.xaxis.axis_label = None
Finally, having the tool bars for each plot is unnecessary and introduces clutter. To only show a single toolbar for a set of subplots, we should instead use bokeh.layouts.gridplot(). We use the keyword argument ncols=1 to specify that we want a single column.
bokeh.io.show(bokeh.layouts.gridplot(plots, ncols=1))
Similarly, we can arrange the plots in a 2×2 grid, we could use ncols=2 as our kwarg. Of course, we have to supply an axis label for the plot in the lower left corner.
plots[2].xaxis.axis_label = 'impact force (mN)'
bokeh.io.show(bokeh.layouts.gridplot(plots, ncols=2))
We can also use bokeh.layouts.gridplot() on a 2-dimensional list, including with blank spaces. To demonstrate, I'll leave out the contact pressure plot.
# Make 2D list of plots (put None for no plot)
plots = [[plots[0], plots[1]],
[plots[2], None]]
# Show using gridplot withou an ncols kwarg
bokeh.io.show(bokeh.layouts.gridplot(plots))
Plotting the distribution of impact forces¶
What if we just want to plot the distribution of impact forces? There are several ways we could do this. Perhaps the most familiar one is to plot them as a histogram. To do that with Bokeh, we can first compute the edges and heights of the bars of the histogram using Numpy, and then add them to the plot using the quad() method of Bokeh figures, which plots filled rectangles.
# Compute the histogram
heights, edges = np.histogram(df['impact force (mN)'])
# Set up the plot
p = bokeh.plotting.figure(plot_height=300,
plot_width=500,
x_axis_label='impact force (mN)',
y_axis_label='count')
# Generate the histogram
p.quad(top=heights, bottom=0, left=edges[:-1], right=edges[1:])
bokeh.io.show(p)
At first glance, there seems to be some bimodality. We could start playing around with the number of bins to look for closely, but I will not bother to do that here. In general, I contend that a histogram is rarely the best way to plot the distribution of repeated measurements. Instead, an ECDF is better....
Empirical cumulative distribution functions (ECDFs)¶
While informative in this case, I generally frown on using histograms to display distributions of measurements. The primary reason for this is binning bias. To create a histogram, we necessarily have to consider not the exact values of measurements, but we must place these measurements in bins. We do not plot all of the data, and the choice of bins can change what you infer from the plot.
Instead, we should look at the empirical cumulative distribution function, or ECDF. The ECDF evaluated at $x$ for a set of measurements is defined as
ECDF(x) = fraction of measurements ≤ x.
You will write a function in your first homework to generate x and y values for ECDFs. Here, I show the image of a Bokeh plot of the ECDF.
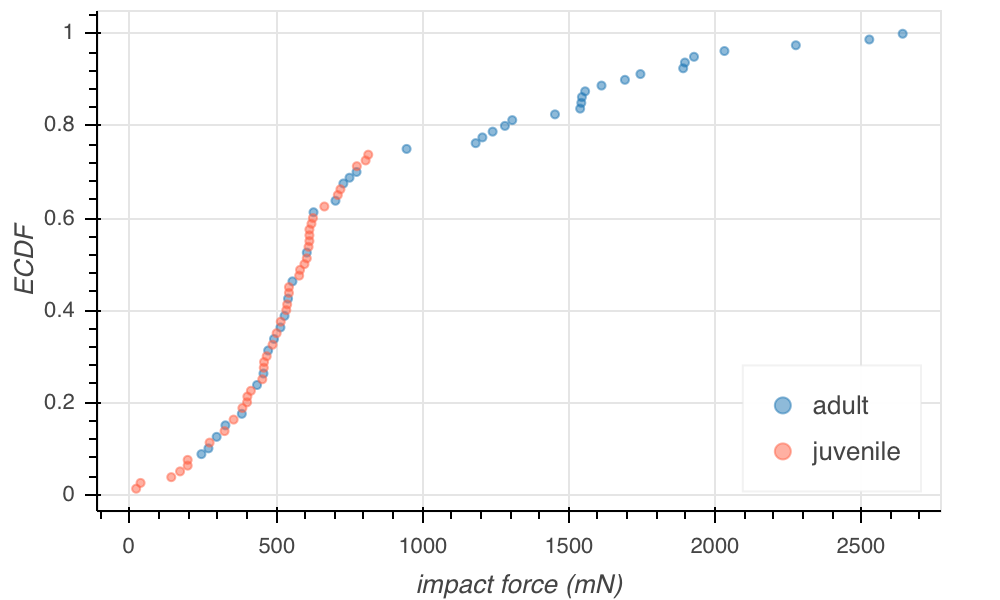
The x value for each point on the plot is the measured result. The y value is the fraction of measurements in the complete data set that were less than or equal to the x value. Importantly, every data point is plotted. There is no binning bias. The ECDF contains all of the empirical information about the distribution, and we can infer bimodality from inflection points in the ECDF.
Note that I have also color coded the data points by whether the frog was a juvenile or adult. Indeed, the bimodality we saw falls along the lines of age of the frog, though there do appear to be some low-impact strikes from some lazy adults here and there.
Conclusions¶
In this tutorial, we have learned how to load data from CSV files into Pandas DataFrames. DataFrames are useful objects for looking at data from many angles. We have just touched the tip of the iceberg of the functionality of Pandas. You will slowly gain more familiarity with its features as you use it and ask Google how to do tasks you have in mind.
And you now know not to make histograms. Make ECDFS.